Fragmentation Analyzer
- What is Fragmentation Analyzer?
- Supported Input Data
- Downloading and Installing
- Tutorial
- Importing Data
- Supported Text File Format
- Support
- Screenshots
Fragmentation Analyzer Publications:
- Barsnes et al: Proteomics 2010 Mar;10(5):1087-90.
- If you use Fragmentation Analyzer as part of a paper, please include the reference above.
NEW! For further analysis of proteomics data we also recommend checking out SearchGUI and PeptideShaker.
What is Fragmentation Analyzer?
Fragmentation Analyzer is a tool for analyzing MS/MS fragmentation data. Currently nine different analysis types are supported:
- Spectra Visualization - visualize the MS/MS spectra with fragment ion annotation, zooming and manual de-novo-sequencing.
- Intensity Box Plots - analyze intensity variation for a set of identification of the same peptide.
- Mass Error Scatter Plots - visualize the mass error spread in a set of selected identifications/spectra.
- Mass Error Bubble Plots - same as Mass Error Scatter Plots but with peak intensities added.
- Mass Error Box Plot - analyze the variation in mass errors.
- Fragment Ion Probability Plot - analyze the probability of observing specific fragment ions.
- Fragment Ion Heat Map - compare fragmentations using heat maps of correlation data.
- Intensity Correlation - detect how the intensity correlates with the spread in intensity.
- Intensity Meta Plots - further analyze the variations in intensity.
See the screenshots for examples of the tool in action.
Fragmentation Analyzer has been developed by Harald Barsnes (University of Bergen, Norway) under the guidance of Prof. Dr. Lennart Martens.
Supported Input Data
Fragmentation Analyzer currently supports three input formats:
In addition any files that can be converted to the supported text file format supported text file format can be used. (See the importing data section for details on how to import data.)
Downloading and Installing
Requirements
- Fragmentation Analyzer requires Java 1.5 (or newer), which you can download for free here. Most modern computers will have Java installed already, so first try to follow the instructions below to download and run Fragmentation Analyzer before downloading Java. You only need the JRE version (and not the JDK version) to run Fragmentation Analyzer.
- Unless your data is stored in, or can be converted to, one of the supported data formats, the software will unfortunately not be of much help.
- The current version has been tested on Windows XP, Windows Vista, Mac and Linux, but should also work on other platforms, as long as Java 1.5 (or newer) is installed. If you come across problems on your platform please let us know.
Downloading
Download the latest version of FragmentationAnalyzer_X.Y.zip (where X and Y represent the version of the software) here. Unzipping the file, creates the following directory structure:
FragmentationAnalyzer_X.Y
FragmentationAnalyzer-X.Y.jar
README.txt
Properties (folder)
DataSets (folder)
lib (folder)
To start the software, simply double-click the file named FragmentationAnalyzer-X.Y.jar.
If this fails, try to download and install Java 1.5 or above, as explained in the previous section. (The program can also be started from the command line using the following command: java -jar FragmentationAnalyzer-X.Y.jar.) A Fragmentation Analyzer icon that can be used for shortcuts etc linking to the program is included in the zip file.
An example data set is included in the DataSet folder.
Troubleshooting
-
Does Not Start - If nothing happens when double clicking the Fragmentation Analyzer jar file, the most likely cause is that you don’t have Java installed. Download the latest version of Java here and try again. (You only need the JRE version (and not the JDK version) to run Fragmentation Analyzer.)
-
Does Not Start II - If Fragmentation Analyzer fails during start-up and you get the “Fragmentation Analyzer - Startup Failed” message, a file called
fragmentation_analyzer.logwill be created in your home directory. In this file you will find detailed information about why the program was not able to start. -
Does Not Start III - The most likely reason for the “Fragmentation Analyzer - Startup Failed” message is that you have installed Fragmentation Analyzer in a path containing special characters, i.e.
[,%, æ, ø, å, etc. Move the converter to a different folder or rename the folder(s) causing the problem and try again. (On Linux Fragmentation Analyzer also has to be run from a path not containing spaces). -
Does Not Start IV - Another reason for the “Fragmentation Analyzer - Startup Failed” message could be that you have set the upper memory limit higher than your computer can handle (see points below).
-
General Error Diagnosis - In the
Propertiesfolder (see section above for the folder structure), there is a file calledErrorLog.txt. This file contains transcripts of any errors that the application has encountered, and can be very useful in diagnosing your problem. -
Memory Issues - Big datasets can require a lot of memory. If the software unexpectedly fails on a big data set, and the software mentions that it ran out of memory, you should try to give the program more memory. This can be done by editing the
JavaOptions.txtfile in thePropertiesfolder (see section above for the folder structure). In this file, change the-Xmx768Moption to a higher number (e.g.,-Xmx1500Mfor a maximum of appr. 1.5GB of memory). Please note that on a 32-bit operating system you can not increase this value beyond 2000M. -
Problem Not Solved? Or Problem Not In List Above? - See Support.
Tutorial
See Tutorial page in the wiki section.
Importing Data
Data can be imported from three different sources:
ms_lims
For ms_lims one logs on to the ms_lims database via a dialog in the tool using ones normal login details. When connected all the required details about the identifications will be downloaded, while some details, e.g., the fragment ion information, is not downloaded but extracted when needed. The database connection will therefore be required during the use of the tool. Please note that depending on the size of the database the process of importing data from ms_lims might take a while. However, the progress of the import will be monitored closely and presented to the user.
Mascot Dat Files
When importing Mascot dat files one simply selects the set of dat files to import and select the Mascot confidence level to use for the identifications. Only identifications above the selected confidence will be imported.
OMSSA OMX Files
Importing OMSSA omx files is done in the same way as for Mascot dat files (except for the setting of the Mascot confidence level of course). However, the instrument name is not included in the omx file and has to be provided manually by the user for each imported file. Also note that the omx file includes very little details about the amino acid modifications, only a number <1>, <2> etc. The OMMSA installation folder (containing the mods.xml and usermods.xml files) therefore also has to be provided.
Supported Text File Format
When a data set is imported into Fragmentation Analyzer it is divided into three parts:
- identifications.txt - details about the peptide identifications
- fragmentIons.txt - details about the fragment ions
- a folder of pkl files - details about the spectra
identifications.txt
For ms_lims data only the identifications.txt file is created. The remaining information is extracted from the database when needed. However, a file called ‘ms_lims.prop’ is also created containing information about the database used.
identifications.txt is a tab separated text file where the first line includes the number of lines in the file, i.e., the number of identifications. The rest of the file consists of one row per identification with the following elements:
- a unique identification index - [Integer]
- the identified peptide sequence (without terminals and modifications) - [String]
- example:
EITALAPSTMK - the modified peptide sequence (with terminals and modifications) - [String]
- example:
NH2-EITALAPSTM<Mox>K-COOH - peptide precursor charge - [Integer]
- name of instrument used - [String]
- name of corresponding pkl spectrum file (see below) - [String]
- spectrum file id (if in ms_lims database) - [Integer]
- the total intensity of all the peaks in the spectrum - [Double]
- original file name (a reference to the original spectrum file) - [String]
Either spectrum file name or spectrum id has to be provided, but the other can be set to “null”.
fragmentIons.txt
fragmentIons.txt is also a tab separated consisting of one row per fragment ion with the following elements:
- a unique fragment ion index - [Integer]
- identification index (a reference to the identification the fragment comes from - [Integer]
- fragment ion type, e.g., y2, b3-H20 etc. - [String]
- fragment ion m/z value - [Double]
- fragment ion intensity - [Double]
- fragment ion number, e.g., y2 has the fragment ion number 2 - [Double]
- fragment ion mass error (the (absolute) distance between the theoretical and the experimental mass of the fragment ion (experimental mass - theoretical mass)) - [Double]
The following fragment ion type names are recommended and will result in the best integration with the tool:
- Standard fragment ions, singly charged: b1, b2, y1, y2, etc.
- Neutral loss ions: y[5]-H2O, y[4]-NH3, etc.
- Doubly charged fragment ions: b[2]++, b[4]++-NH3, etc.
- Precursor ions: Prec, Prec-H2O 2+, etc.
- Immonium ions: iA, iC, etc.
Spectra Folder
For non-ms_lims data sets the spectra are stored as pkl files in a folder called ‘spectra’. One file per spectrum. The first line in each file contains the precursor m/z, intensity and charge. Next follows one line per peak in the spectrum with the m/z and intensity values.
For more details see the example data set or the source code.
Support
For questions or additional help, feel free to contact the authors. If appropriate please include a (preferably zipped) copy of the ErrorLog.txt file from the Properties folder.
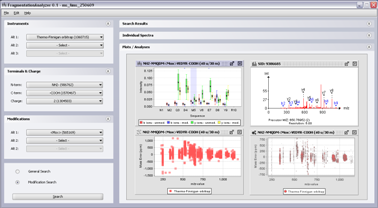
Screenshots
(Click on the screenshot to see the full size version)
Screenshot 1: Main interface of Fragmentation Analyzer.
(Click on the screenshot to see the full size version)
Screenshot 2: Main interface of Fragmentation Analyzer with plots.
(Click on the screenshot to see the full size version)
Screenshot 3: Boxplot of fragment ions for modified vs unmodified peptides.
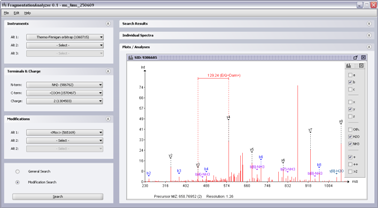
(Click on the screenshot to see the full size version)
Screenshot 4: MS/MS spectra with fragment ion annotation.
(Click on the screenshot to see the full size version)
Screenshot 5: Mass Error Bubble Plot. Size of bubble relative to intensity. Color coding: instrument type.
(Click on the screenshot to see the full size version)
Screenshot 6: Mass Error Bubble Plot. Size of bubble relative to intensity. Color coding fragment ion type.
(Click on the screenshot to see the full size version)
Screenshot 7: Fragment Ion Probability Plot.
(Click on the screenshot to see the full size version)
Screenshot 8: Fragment Ion Heat Map - analyze the correlation of different fragmentation patterns.