MAPPI-DAT
Project Description
MAPPI-DAT (Mappit microArray Protein Protein Interaction- Data management & Analysis Tool) is an automated high-throughput data management and analysis system for microarray-MAPPIT system. MAPPI-DAT is capable of processing many thousand data points for each experiment, and comprising a data storage system that stores the experimental data in a structured way.
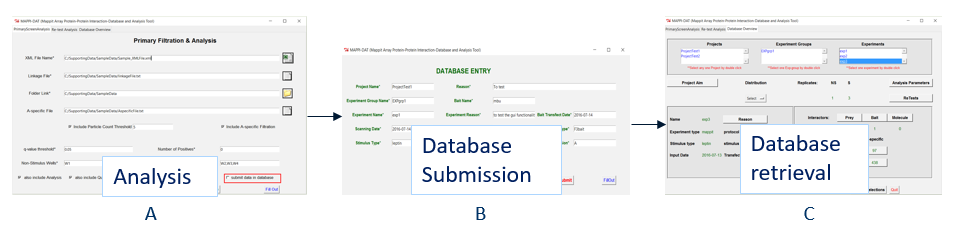
MAPPI-DAT application include graphical user interface. The analysis system is comprising of three steps: normalization, analysis and post filtration. The management system contains a back-end relational database and retrieval system, allowing user to store and download different projects using MAPPI-DAT interface with three main divisions for analysis, database submission and retrieval.
Citation
“MAPPI-DAT: data management and analysis for protein-protein interaction data from the high-throughput MAPPIT cell microarray platform” Surya Gupta, Veronic De Puysseleyr, José Van der Heyden, Davy Maddelein, Irma Lemmens, Sam Lievens, Sven Degroeve, Jan Tavernier, and Lennart Martens. Bioinformatics 2017; 33 (9): 1424-1425, PMID: 28104627
Downloads
Python project: MAPPI-DAT project can be downloaded from MAPPI-DAT.
Windows stand-alone files: window compatible .exe files can be downloaded from:
database schema: MySQL schema for the MAPPI-DAT can be downloaded from here
Usage
See the manual for further information about how to use the tool.
Sample data can be downloaded from here
Project Support
MAPPI-DAT project is supported by:
| Compomics | VIB | Ghent University |
|---|---|---|
 |
 |
 |