Usage And Examples
Traml Standard
TraML is a new standardized format for encoding transition lists and associated metadata developed by the Proteomics Standards Initiative.
TraML builds on the same design concepts that were used for mzML and mzIdentML. Like these formats previously developed for different data types, TraML is based on Extensible Markup Language (XML) and can be parsed and validated for structural correctness with a large variety of industry-standard tools. As with the other PSI formats, most of the metadata in the TraML file are encoded with the use of controlled vocabulary (CV) terms. These terms are all included in the PSI-MS CV, the same CV used by mzML and mzIdentML, and actively maintained by the PSI MSSWG.
jTraML API
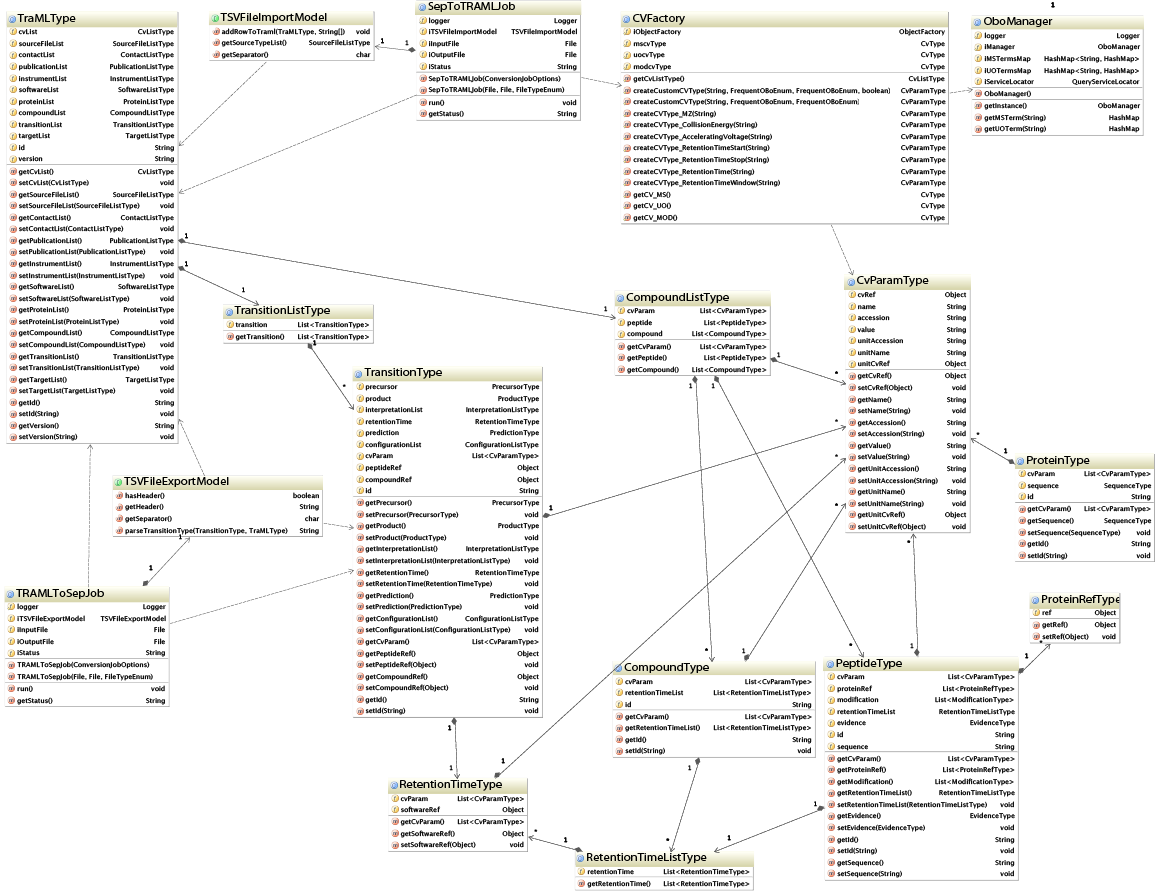
Reduced UML diagram
The TraMLType is the root element of the TraML file. Central in this diagram are the TransitionType and CompoundType/PeptideType elements which represent all transitions that need to be analyzed in a SRM experiment. Note that a PeptideType in term references a ProteinType, and also links to a RetentionTimeType element in order to define expected chromatographic elution times.
Aside from these JAXB generated classes, the reduced UML diagram highlights jTraML convenience classes such as the CVFactory and the OboManager, both working together to detail each of the TraML elements with Controlled Vocabulary based Parameters.
Furthermore, the TSVFileImportModel and TSVFileExportModel declare a conversion interface for generating TraML or TSV files respectively. The SepToTRAMLJob and the TRAMLToSepJob in turn make use of these models for threaded conversion of SRM input files.
Conversion Methods
ABI csv
example transition:
# 564.9618,663.4081,10,LSTADPADASTIYAVVV.O95866.O95866-3.O95866-5.3y6,29.3
- 564.9618 = precursor ion m/z (Q1)
- 663.4081 = product ion m/z (Q3)
- 10 = ???
- LSTADPADASTIYAVVV = peptide sequence
- O95866 = protein accession (A)
- O95866-3 = protein accession (B)
- O95866-5 = protein accession (C)
- 3y6 = precursor charge 3+, fragment y6
- 29.3 = centroid retention time (min)
Thermo csv
example transition:
# Q1,Q3,CE,Start time (min),Stop time (min),Polarity,Trigger,Reaction category,Name
# 651.8366,790.4038,25.5,18.61,28.61,1,1.00E+04,0,AAELQTGLETNR.2y7-1
- 651.8366 = precursor ion m/z (Q1)
- 790.4038 = product ion m/z (Q3)
- 25.5 = collision energy
- 18.61 = start time (min)
- 28.61 = stop time (min)
- 1 = polarity
- 1.00E+04 = trigger
- 0 = reaction category
- AAELQTGLETNR.2y7-1 = name
Agilent tsv
example transition:
# Dynamic MRM
# Compound NameISTD?Precursor IonMS1 ResProduct IonMS2 ResFragmentorCollision EnergyCell Accelerator VoltageRet Time (min)Delta Ret TimePolarity
# CSASVLPVDVQTLNSSGPPFGK.2y16-1FALSE1130.5681Wide1642.8233Unit12539.8542.355.00Positive
- CSASVLPVDVQTLNSSGPPFGK.2y16-1 = name
- FALSE = ISTD (?)
- 1130.5681 = precursor ion m/z (Q1)
- Wide = MS1 resolution
- 1642.8233 = product ion m/z (Q3)
- Unit = MS2 resolution
- 125 = fragmentor
- 39.8 = collision energy
- 5 = cell accelerator voltage
- 42.35 = centroid retention time
- 5.00 = delta retention time
- Positive = polarity
TraML Converter Command Line
The TraML Converter is the starter class in the jtraml-core library. So executing the following statement in the command line will launch the jTraML converter.
java -jar jtraml-core-x.x.jar
If no options are provided, then the command line usage will be displayed in the prompt.
TraMLConverter
----------------------
INFO
----------------------
The TraML converter command line tool takes an input file and an input type and generates an output file.
If the input type is a .TraML file, then the converter generates be a .TSV file.
Otherwise, if the input type is a .TSV file, then the converter generates a .TraML file.
----------------------
OPTIONS
----------------------
Core options:
- exporttype <arg>The available file types:<TraML><thermo_csv><agilent_tsv><abi_csv>
- importtype <arg>The available file types:<TraML><thermo_csv><agilent_tsv><abi_csv>
- input <arg> The transition input file
- output <arg> The converted transition output file
- rtdelta <arg> This delta retention time (minutes) is used when appropriate (cfr. Wiki)
- rtshift <arg> The retention time shift (minutes) value is used to added to the present retention times. Can be positive or negative. (cfr. Wiki)
Constant options:
- fragmentor <arg>constant - agilent - LC/MS value '125'
- istd <arg> constant - agilent - internal standard - TRUE/FALSE
- qtrapcol3 <arg> constant - abi - tsv column 3 variable '10'
- rcategory <arg> constant - thermo - reaction category (iSRM) - Set 0 for primaries, or 1 for secondaries.
- resms1 <arg> constant - agilent - MS1 resolution - 'Unit' (0.7AMU) - 'Wide' (1.2AMU) - 'Widest' (2.5AMU)
- resms2 <arg> constant - agilent - MS2 resolution - 'Unit' (0.7AMU) - 'Wide' (1.2AMU) - 'Widest' (2.5AMU)
- trigger <arg> constant - thermo - set 1 to trigger recording of full MS/MS spectra.
TraML Converter Web
http://iomics.ugent.be/jtraml
TraML Converter Retention Time Conversion Scenarios

Using jTraML as a Maven dependency
MascotDatfile is hosted at our public maven repository.
Add the following code into your pom.xml file:
Repository
<repositories>
<!-- Compomics Genesis Maven 2 repository -->
<repository>
<id>genesis-maven2-repository</id>
<name>Genesis maven2 repository</name>
<url>http://genesis.UGent.be/maven2</url>
<layout>default</layout>
</repository>
</repositories>
Dependency
<dependencies>
<dependency>
<groupId>com.compomics.jtraml</groupId>
<artifactId>jtraml-core</artifactId>
<version>X.Y.Z</version>
<type>jar</type>
</dependency>
</dependencies>
Update the version number (X.Y.Z) to latest released version.
Note that the repository can be manually accessed to download the src or javadocs.